- News
PNAS│Hongxia Wang's Team Reveals Metabolic Serum Fingerprinting of Breast Cancer
Link of original article:
https://mp.weixin.qq.com/s/rWZyvuapyafJjUbXfprzuw
Breast cancer is located at the top of the incidence of malignant tumors in women, and efficient analytical tools are crucial for early and effective diagnosis and prognosis prediction of breast cancer. Currently, clinical practice still relies on conventional methods of histopathological classification and imaging tests, such as mammography, magnetic resonance imaging and ultrasonography, which have a lag in early diagnosis and detection of recurrent metastases.

On March 19, 2022 PNAS published online the research results of Wang Hongxia's team from Shanghai General Hospital of Shanghai Jiaotong University School of Medicine on high performance breast cancer metabolic fingerprinting for breast cancer diagnosis and prognosis prediction, entitled: Diagnosis and prognosis of breast cancer by high-performance serum metabolic fingerprints, using high-throughput NPELDI-MS technology instead of traditional mass spectrometry assays, and resolving the serum metabolic fingerprints of breast cancer patients, benign breast diseases and healthy population through a large sample clinical cohort study.
The analysis of the metabolic profile of breast cancer development and recurrent metastasis, which is accompanied by alterations in the biological phenotype and metabolic energy supply of cancer cells, can help reveal the real-time status of disease progression. In this study, serum metabolic fingerprints (SMFs) of breast cancer patients, benign breast disease and healthy populations were resolved in a large clinical cohort study using high-throughput NPELDI-MS technology instead of traditional mass spectrometry assays. The study showed that SMFs-based metabolomic information obtained from breast cancer was significantly different between breast cancer patients and non-breast cancer individuals; SMFs performed well in the early diagnosis of breast cancer and non-breast cancer patients (AUC 0.948, 95% CI: 0.922-0.973, accuracy 88.8%, sensitivity 88.9%, specificity 88.8%) that showed good prospects for clinical application.
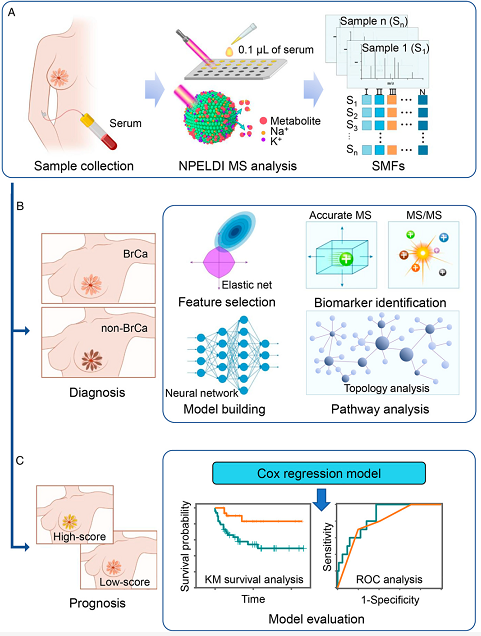
Further, the team used Cox regression model to construct a 4-metabolite based metabolic prognostic scoring system (MP-score), and ROC analysis showed that the 4-metabolite based MP-score showed high sensitivity and specificity for prognosis and recurrence metastasis monitoring of breast cancer patients in both the test and validation cohorts, significantly outperforming the traditional TNM staging system. The metabolic prognostic scoring system established by SMFs was effective in predicting patient prognosis and survival (p < 0.005). The fingerprinting and detection technology has the advantages of good reproducibility, rapid analysis capability (only 30 seconds per sample) and small sample consumption (only 100 nL of serum), which facilitates the promotion of large-scale clinical applications in the future and makes high-performance rapid blood tests possible in the clinic.
For the identification of metabolic biomarkers and pathway co-analysis, the team used stepwise screening model analysis to identify seven metabolic molecules from SMFs data matrix that were significantly different in breast cancer patients, and validated by FT-ICR-MS or TOF-MS to reveal that these seven metabolites were L-glyceric acid (GA), nicotinamide (NAM), histamine (His), uracil (Ura) , thymine (Thy), 3,4-dihydroxybenzylamine (DB) and dehydroxyphenylalanine (DP). Among them, His, Ura, Thy, DB and DP expression were down-regulated in breast cancer patients (p < 0.05), while GA and NAM expression were up-regulated (p < 0.001) compared to benign breast disease and healthy populations. Further pathway enrichment analysis identified the biological relevance and metabolic signaling pathways of these seven metabolites involving i) pyrimidine metabolism, ii) nicotinic acid and nicotinamide metabolism, and iii) histidine metabolic pathway, respectively. Pyrimidine metabolism reflects adaptive metabolic reprogramming upregulated transcriptional activity due to cancer cell proliferation; nicotinic acid and nicotinamide metabolism represents a high conversion rate of nicotinamide adenine dinucleotide (NAD+) in cancer cells in response to high cancer cell proliferation rate and DNA synthesis; histidine metabolism is closely associated with inflammation and immune response regulation.
This work provides a new target for the follow-up of metabolism-related mechanisms in breast cancer based on metabolomic studies, while the construction of serum metabolic fingerprints provides an effective tool for clinical hematological testing and highlights metabolic features as potential diagnostic and prognostic factors for the disease, which can be extended to other tumors in addition to breast cancer.
The study was conducted by Hongxia Wang, Director of Shanghai First People's Hospital, in collaboration with Kun Qian, a researcher at the School of Biomedical Engineering, Shanghai Jiao Tong University, with Xue-Da Huang, M.S., Shao-Qian Du, M.S., and Jun Liu, Associate Chief Physician, as co-first authors.
